library(tibble)
v1 <- c(1:5)
t1 <- tibble("Number"=v1,"Square"=v1**2)
print(t1)# A tibble: 5 × 2
Number Square
<int> <dbl>
1 1 1
2 2 4
3 3 9
4 4 16
5 5 25The Tidyverse is a collection of R packages developed with an aim to effectively perform data analysis and visualization. The Tidyverse packages are designed to work together seamlessly, and they provide a consistent set of tools for data manipulation, visualization, and modeling. Run install.packages("tidyverse") to install this package. The table below list the core packages available in tidyverse. There are some additional packages as well in addition to these core packages. You can either load individual packages as required or run library(tidyverse) to load all these core packages in one go.
| Package | Description |
|---|---|
| dplyr | Data manipulation and transformation. |
| ggplot2 | Data visualization. |
| readr | Reading data from a variety of sources. |
| tibble | Data structure that is optimized for data science. |
| purrr | Functional programming in R. |
| stringr | String manipulation. |
| forcats | Factor variables. |
| lubridate | Date and time manipulation. |
| tidyr | For data tidying. |
A modern, and often used, variant of a data.frame is a tibble. Tibbles are data frames with some differences in the implementation details making it easier to work with (large) data. After importing the tibble library, the syntax for creating a tibble is same as that for creating a dataframe.
library(tibble)
v1 <- c(1:5)
t1 <- tibble("Number"=v1,"Square"=v1**2)
print(t1)# A tibble: 5 × 2
Number Square
<int> <dbl>
1 1 1
2 2 4
3 3 9
4 4 16
5 5 25An existing data.frame can be converted to a tibble using as_tibble function.
df2 <- data.frame(matrix(c(1:15), nrow = 5, ncol = 3))
t2 <- as_tibble(df2)
print(t2)# A tibble: 5 × 3
X1 X2 X3
<int> <int> <int>
1 1 6 11
2 2 7 12
3 3 8 13
4 4 9 14
5 5 10 15data.frame and tibble| Feature | data.frame | tibble |
|---|---|---|
| Column names | An valid R variable name |
Any alphanumeric text including spaces |
| Row names | Yes | No |
| Subsetting | may return a data.frame or a vector |
Always returns a tibble |
$ operator |
Partially matching can be done | Only exact matches are allowed |
| Printing | Prints entire dataframe | Prints only first 10 rows. |
For more information on these difference you may run vignette("tibble") on the R console.
dplyr libraryIn R, dataframe is a data structure that arranges a data in the form of rows and columns. This can be considered as an equivalent to the familiar spreadsheet kind of arrangement of data. The dplyr library one of the standard library in R for working with dataframes. This library has several functions that are useful for performing related to data analysis. Below we’ll see some of these functions in action using the iris dataset. To load the dplyr library just run the following command.
library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionNext, we’ll read the iris dataset as a tibble.
iris <- as_tibble(iris)
iris# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
7 4.6 3.4 1.4 0.3 setosa
8 5 3.4 1.5 0.2 setosa
9 4.4 2.9 1.4 0.2 setosa
10 4.9 3.1 1.5 0.1 setosa
# ℹ 140 more rowsThe select function is used to select one or more columns from a dataframe. The selection can be made using column names or by specifying a range of columns or by the datatype of the column. When selecting with a range, the name of the starting and the ending column needs to be specified with colon : operator. All the columns within this column range are selected. The code below shows two examples of the select function – first to select the Sepal.Width and Species columns and second to select all the columns starting form the Sepal.Width to Species columns. Note that the column names for the iris dataframe are Sepal.Length, Sepal.Width, Petal.Length, Petal.Width, and Species.
select(iris, Sepal.Width,Species)# A tibble: 150 × 2
Sepal.Width Species
<dbl> <fct>
1 3.5 setosa
2 3 setosa
3 3.2 setosa
4 3.1 setosa
5 3.6 setosa
6 3.9 setosa
7 3.4 setosa
8 3.4 setosa
9 2.9 setosa
10 3.1 setosa
# ℹ 140 more rowsselect(iris, Sepal.Width:Species)# A tibble: 150 × 4
Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <fct>
1 3.5 1.4 0.2 setosa
2 3 1.4 0.2 setosa
3 3.2 1.3 0.2 setosa
4 3.1 1.5 0.2 setosa
5 3.6 1.4 0.2 setosa
6 3.9 1.7 0.4 setosa
7 3.4 1.4 0.3 setosa
8 3.4 1.5 0.2 setosa
9 2.9 1.4 0.2 setosa
10 3.1 1.5 0.1 setosa
# ℹ 140 more rowsThe selections can be made using the Boolean operators (&, |, and !)as well. In addition, the selections can be combined using the c function.
select(iris, !Sepal.Width, Species)# A tibble: 150 × 4
Sepal.Length Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <fct>
1 5.1 1.4 0.2 setosa
2 4.9 1.4 0.2 setosa
3 4.7 1.3 0.2 setosa
4 4.6 1.5 0.2 setosa
5 5 1.4 0.2 setosa
6 5.4 1.7 0.4 setosa
7 4.6 1.4 0.3 setosa
8 5 1.5 0.2 setosa
9 4.4 1.4 0.2 setosa
10 4.9 1.5 0.1 setosa
# ℹ 140 more rowsselect(iris, !(Sepal.Width | Petal.Width), Species)# A tibble: 150 × 3
Sepal.Length Petal.Length Species
<dbl> <dbl> <fct>
1 5.1 1.4 setosa
2 4.9 1.4 setosa
3 4.7 1.3 setosa
4 4.6 1.5 setosa
5 5 1.4 setosa
6 5.4 1.7 setosa
7 4.6 1.4 setosa
8 5 1.5 setosa
9 4.4 1.4 setosa
10 4.9 1.5 setosa
# ℹ 140 more rowsIn addition to selecting based on full names of the columns, selections can also be made by performing pattern matching in the column names using the helper functions - starts_with, ends_with, contains, matches (regular expression), and num_range (to match a numerical range).
select(iris, starts_with("S"))# A tibble: 150 × 3
Sepal.Length Sepal.Width Species
<dbl> <dbl> <fct>
1 5.1 3.5 setosa
2 4.9 3 setosa
3 4.7 3.2 setosa
4 4.6 3.1 setosa
5 5 3.6 setosa
6 5.4 3.9 setosa
7 4.6 3.4 setosa
8 5 3.4 setosa
9 4.4 2.9 setosa
10 4.9 3.1 setosa
# ℹ 140 more rowsselect(iris, contains("Petal"), Species)# A tibble: 150 × 3
Petal.Length Petal.Width Species
<dbl> <dbl> <fct>
1 1.4 0.2 setosa
2 1.4 0.2 setosa
3 1.3 0.2 setosa
4 1.5 0.2 setosa
5 1.4 0.2 setosa
6 1.7 0.4 setosa
7 1.4 0.3 setosa
8 1.5 0.2 setosa
9 1.4 0.2 setosa
10 1.5 0.1 setosa
# ℹ 140 more rowsThere is yet another set of helper functions to make a selection e.g. last_col function selects, as the name suggests, the last column. This function takes an optional keyword argument offset to change the counting for the last column i.e. last_col(1) will select the second last column. Similarly, the everything function can be used to select everything (duh!). At first this function might not seem very useful but it comes handy when we want to reorder the occurence of the columns in the dataframe. E.g., in the select function, we can first specify the columns that we want in the beginning and then specify everthing to retain the remaining columns.
select(iris, last_col())# A tibble: 150 × 1
Species
<fct>
1 setosa
2 setosa
3 setosa
4 setosa
5 setosa
6 setosa
7 setosa
8 setosa
9 setosa
10 setosa
# ℹ 140 more rowsselect(iris, Species, everything())# A tibble: 150 × 5
Species Sepal.Length Sepal.Width Petal.Length Petal.Width
<fct> <dbl> <dbl> <dbl> <dbl>
1 setosa 5.1 3.5 1.4 0.2
2 setosa 4.9 3 1.4 0.2
3 setosa 4.7 3.2 1.3 0.2
4 setosa 4.6 3.1 1.5 0.2
5 setosa 5 3.6 1.4 0.2
6 setosa 5.4 3.9 1.7 0.4
7 setosa 4.6 3.4 1.4 0.3
8 setosa 5 3.4 1.5 0.2
9 setosa 4.4 2.9 1.4 0.2
10 setosa 4.9 3.1 1.5 0.1
# ℹ 140 more rowsThe filter function is used to create subset of the dataframe based on certain condition(s). When specifying multiple conditions, Boolean operators can be used (&, |, !). In addition, functions such as between, near, etc. can also be used. A simple use of filter can be to select all the rows in which a particular column has a given value. E.g., to select all the rows in iris where the Species column is setosa, we need to specify Species==setosa as an argument to the filter function. This would result in dataframe with 50 rows. Let’s say we want to select rows where the species name is setosa the Petal.Length is greater than 1.5. In this case the filter function would have two conditions with Boolean and (&). This would give a dataframe with 13 rows.
filter(iris, Species == "setosa")# A tibble: 50 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
7 4.6 3.4 1.4 0.3 setosa
8 5 3.4 1.5 0.2 setosa
9 4.4 2.9 1.4 0.2 setosa
10 4.9 3.1 1.5 0.1 setosa
# ℹ 40 more rowsfilter(iris, Species=='setosa' & Petal.Length>1.5)# A tibble: 13 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5.4 3.9 1.7 0.4 setosa
2 4.8 3.4 1.6 0.2 setosa
3 5.7 3.8 1.7 0.3 setosa
4 5.4 3.4 1.7 0.2 setosa
5 5.1 3.3 1.7 0.5 setosa
6 4.8 3.4 1.9 0.2 setosa
7 5 3 1.6 0.2 setosa
8 5 3.4 1.6 0.4 setosa
9 4.7 3.2 1.6 0.2 setosa
10 4.8 3.1 1.6 0.2 setosa
11 5 3.5 1.6 0.6 setosa
12 5.1 3.8 1.9 0.4 setosa
13 5.1 3.8 1.6 0.2 setosa There is another way of combining different filter/selection operations which by using the concept of chaining. In this approach the different functions areas applied one after the other such that the output of one operation is the input for the next – hence the name chaining. The different commands are chained using the %>% operator. In this approach, we need to specify that dataframe only in the beginning followed by the set of operations. The output of the chained operation can be redirected (<-) to a variable.
df_out <- iris %>%
filter(Species=='setosa') %>%
filter(Petal.Length>1.5) %>%
select(starts_with("S"))
df_out# A tibble: 13 × 3
Sepal.Length Sepal.Width Species
<dbl> <dbl> <fct>
1 5.4 3.9 setosa
2 4.8 3.4 setosa
3 5.7 3.8 setosa
4 5.4 3.4 setosa
5 5.1 3.3 setosa
6 4.8 3.4 setosa
7 5 3 setosa
8 5 3.4 setosa
9 4.7 3.2 setosa
10 4.8 3.1 setosa
11 5 3.5 setosa
12 5.1 3.8 setosa
13 5.1 3.8 setosa As you can see, this method is much better in terms of readability. We can read the three lines of code above as — take the iris dataframe, select rows where Species is setosa, select rows where Petal.Length is greater than 1.5 and finally select the columns for which the name starts with “S”. Another advantage of chaining is that we can easily add new selections or other operations using the %>% operator. It is because of these reasons that chaining is the most preferred way of applying multiple functions to a dataframe.
Quiz
Using the mtcars data set, write a program to select cars with more than 4 cylinders and more than 4 gears.
as_tibble(mtcars) %>%
filter(cyl > 4) %>%
filter(gear > 4)
## Note that we can use Boolean logic as well.
## filter (cyl > 4 & gear > 4)When there is a need to re-arrange rows in a dataframe based on certain criteria, we can use the arrange function. E.g., this function comes handy to sort a dataframe based on values in a particular column. The code below re-orders the rows in the iris dataframe based on the values in the Petal.Length column. Note that the default sorting is in ascending order for number and alphabetical for strings. The desc (descending) function can be used to change the default behavior.
arrange(iris, Petal.Length)# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 4.6 3.6 1 0.2 setosa
2 4.3 3 1.1 0.1 setosa
3 5.8 4 1.2 0.2 setosa
4 5 3.2 1.2 0.2 setosa
5 4.7 3.2 1.3 0.2 setosa
6 5.4 3.9 1.3 0.4 setosa
7 5.5 3.5 1.3 0.2 setosa
8 4.4 3 1.3 0.2 setosa
9 5 3.5 1.3 0.3 setosa
10 4.5 2.3 1.3 0.3 setosa
# ℹ 140 more rowsiris %>%
arrange(desc(Petal.Length))# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 7.7 2.6 6.9 2.3 virginica
2 7.7 3.8 6.7 2.2 virginica
3 7.7 2.8 6.7 2 virginica
4 7.6 3 6.6 2.1 virginica
5 7.9 3.8 6.4 2 virginica
6 7.3 2.9 6.3 1.8 virginica
7 7.2 3.6 6.1 2.5 virginica
8 7.4 2.8 6.1 1.9 virginica
9 7.7 3 6.1 2.3 virginica
10 6.3 3.3 6 2.5 virginica
# ℹ 140 more rowsTo rename a column, use rename with an argument new name = old name. E.g. to rename the Species column of the iris dataframe to iris.species the following code can be used.
iris %>%
rename(iris.species = Species)# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width iris.species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
7 4.6 3.4 1.4 0.3 setosa
8 5 3.4 1.5 0.2 setosa
9 4.4 2.9 1.4 0.2 setosa
10 4.9 3.1 1.5 0.1 setosa
# ℹ 140 more rowsOne of the common tasks when doing data analysis is to work with a subset of the data. For instance, in case of the iris data set let’s say we want to work with one of the species (e.g. setosa) and would like to know the mean petal length for this species. This can be easily done by first selecting the rows where the species is setosa and then calculating the mean for the petal length column. The code below uses the pull function to extract a column as a numeric vector and then uses the mean function to calculate the aveage petal length for setosa. This analysis can be extended to other species in the dataset as well.
pl <- iris %>%
filter(Species == "setosa") %>%
pull(Petal.Length) %>%
mean()
pl[1] 1.462A better approach to perform this task is to use group_by function to group the observations (rows) by species name and then use the summarize function the calculate mean. The group_by function would internally create sub-set of the dataframe based on the different species name in the Species column. This way we’ll can perform the desired operation on each of the species separately and in one go.
iris %>%
group_by(Species) %>%
summarise(mean(Petal.Length))# A tibble: 3 × 2
Species `mean(Petal.Length)`
<fct> <dbl>
1 setosa 1.46
2 versicolor 4.26
3 virginica 5.55While summarizing we can perform multiple operations on as many columns as we like. The column names of the resulting dataframe can also be specified. The code below calculate the mean and standard deviation of petal length and sepal length columns for each of the three species.
iris %>%
group_by(Species) %>%
summarise(PL_mean = mean(Petal.Length), PL_stdev = sd(Petal.Length),SL_mean = mean(Sepal.Length), SL_stdev = sd(Sepal.Length))# A tibble: 3 × 5
Species PL_mean PL_stdev SL_mean SL_stdev
<fct> <dbl> <dbl> <dbl> <dbl>
1 setosa 1.46 0.174 5.01 0.352
2 versicolor 4.26 0.470 5.94 0.516
3 virginica 5.55 0.552 6.59 0.636When working with dataframe, at times we need to add additional columns to the dataframe which are derived from the data in the existing columns. E.g., in the iris dataframe, let’s say we want to add a column that has sum of Petal.Length and Sepal.Length. This can be easily achieved using the mutate function. By default, the new column is added at the end of the dataframe. The optional keyword arguments for mutate – .before and .after can be used to specify the column before/after which the new column should be added
iris_mod <- iris %>%
mutate(sum1 = Petal.Length + Sepal.Length)
iris_mod# A tibble: 150 × 6
Sepal.Length Sepal.Width Petal.Length Petal.Width Species sum1
<dbl> <dbl> <dbl> <dbl> <fct> <dbl>
1 5.1 3.5 1.4 0.2 setosa 6.5
2 4.9 3 1.4 0.2 setosa 6.3
3 4.7 3.2 1.3 0.2 setosa 6
4 4.6 3.1 1.5 0.2 setosa 6.1
5 5 3.6 1.4 0.2 setosa 6.4
6 5.4 3.9 1.7 0.4 setosa 7.1
7 4.6 3.4 1.4 0.3 setosa 6
8 5 3.4 1.5 0.2 setosa 6.5
9 4.4 2.9 1.4 0.2 setosa 5.8
10 4.9 3.1 1.5 0.1 setosa 6.4
# ℹ 140 more rowsWe can also add customs values in the new column base on some conditions e.g., to add a new column to iris based on whether the sepal length is greater than five or not, the following code can be used. Here, the if_else block checks condition, if it is true then the first value is added to the column otherwise the second value is added to the column. The .after specifies the location where to add the new column.
iris_mod <- iris %>%
mutate(SL_condition = if_else(.$Sepal.Length>=5, "Greater than or equal to 5", "Less than 5"), .after = Sepal.Length)
iris_mod# A tibble: 150 × 6
Sepal.Length SL_condition Sepal.Width Petal.Length Petal.Width Species
<dbl> <chr> <dbl> <dbl> <dbl> <fct>
1 5.1 Greater than or eq… 3.5 1.4 0.2 setosa
2 4.9 Less than 5 3 1.4 0.2 setosa
3 4.7 Less than 5 3.2 1.3 0.2 setosa
4 4.6 Less than 5 3.1 1.5 0.2 setosa
5 5 Greater than or eq… 3.6 1.4 0.2 setosa
6 5.4 Greater than or eq… 3.9 1.7 0.4 setosa
7 4.6 Less than 5 3.4 1.4 0.3 setosa
8 5 Greater than or eq… 3.4 1.5 0.2 setosa
9 4.4 Less than 5 2.9 1.4 0.2 setosa
10 4.9 Less than 5 3.1 1.5 0.1 setosa
# ℹ 140 more rowsThe mutate function can also be used to mutate the data type for column(s). The across function allows the application of transformation to multiple columns.
iris %>%
mutate(across((Sepal.Length:Sepal.Width), as.integer))# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<int> <int> <dbl> <dbl> <fct>
1 5 3 1.4 0.2 setosa
2 4 3 1.4 0.2 setosa
3 4 3 1.3 0.2 setosa
4 4 3 1.5 0.2 setosa
5 5 3 1.4 0.2 setosa
6 5 3 1.7 0.4 setosa
7 4 3 1.4 0.3 setosa
8 5 3 1.5 0.2 setosa
9 4 2 1.4 0.2 setosa
10 4 3 1.5 0.1 setosa
# ℹ 140 more rowsOne of the pre-requisites for effective data analysis is having the data in the right format. And, in tidyverse the right format means the tidy format which imply that each row should correspond to an observation and each column should correspond to a unique variable (or feature).
The pivot_longer function transforms a dataframe in a wide form to a long form by adding rows and reducing the number of columns. For a given column in a dataframe, this function adds two columns in the dataframe - a “name” column that has the original column name as the column values and a “value” column that has the values for that column.
In the example below, we have a dataframe have marks in Maths and Science subjects for four students. There are three columns correponding to the name of the students, marks in maths and marks in science. Now, this dataframe is not a “tidy” dataframe because there are more than one column having same type of information i.e. marks. Although these are marks in different subjects but for this dataframe to be “tidy” there should be only one column for the marks. So, we need to change the layout out this dataframe such that the new layout would have three columns – students name, subject, and marks. This can be achieved using the pivot_longer function. Notice that in the “tidy” layout there will greater number of rows (eight) as compared to the original layout of the dataframe (four).
library(tidyverse)Warning: package 'ggplot2' was built under R version 4.4.3Warning: package 'purrr' was built under R version 4.4.3── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ forcats 1.0.0 ✔ readr 2.1.5
✔ ggplot2 3.5.2 ✔ stringr 1.5.1
✔ lubridate 1.9.4 ✔ tidyr 1.3.1
✔ purrr 1.0.4
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsStudents <- c("Sam", "Mohan", "Mike", "Raj")
maths <- c(60,70,80,90)
science <- c(65,75,85,95)
df1 <- data.frame(cbind(Students, maths, science))
df1$maths <- as.integer(df1$maths)
class(df1$science) <- "integer"
print(df1) Students maths science
1 Sam 60 65
2 Mohan 70 75
3 Mike 80 85
4 Raj 90 95df2 <- df1 %>%
pivot_longer(c("maths", "science"), names_to = "Subject", values_to = "Marks")
print(df2)# A tibble: 8 × 3
Students Subject Marks
<chr> <chr> <int>
1 Sam maths 60
2 Sam science 65
3 Mohan maths 70
4 Mohan science 75
5 Mike maths 80
6 Mike science 85
7 Raj maths 90
8 Raj science 95The advantage of having data in the “tidy” format is that now it would work well with the different functions in the tidyverse. For instance, we can now easily plot the pairwise marks for all the students using ggplot. This would have been a non-trivial exercise had we not used the pivot_longer function with our original dataframe. We learn more abouqt plotting using ggplot in the chapter on Data Visualization
df2 %>%
ggplot() + aes(x=Students, y=Marks, fill=Subject) + geom_col(width=0.5, position = "dodge") + scale_y_continuous() + scale_fill_brewer(palette="Paired")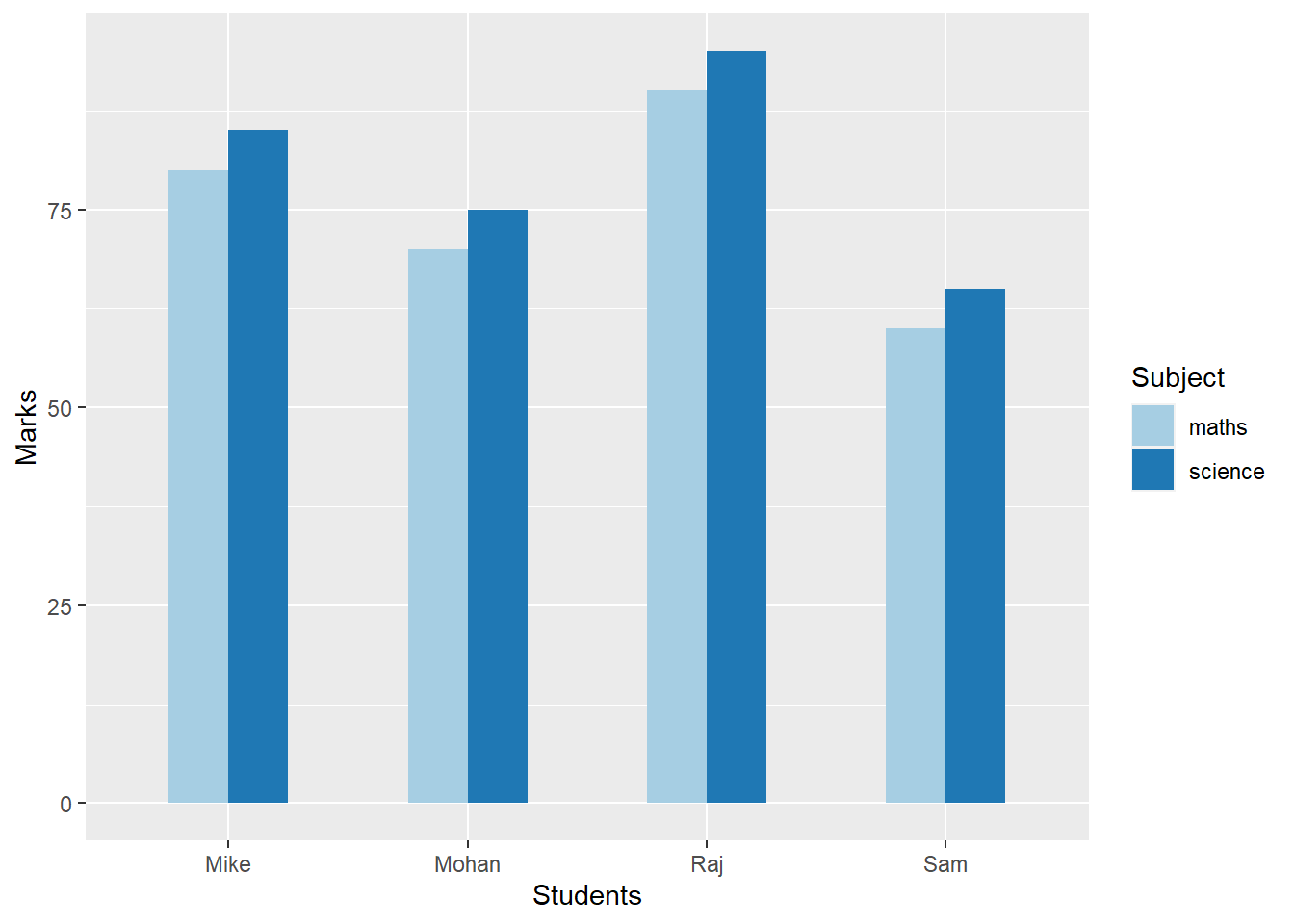
The pivot_wider function, as the name suggests, works in opposite manner to the pivot_longer function i.e. it makes a dataframe wider by adding new columns and decreasing the total number of rows. Let’s look at an example to get a better understanding this function. Suppose we have each student enrolled in two courses from a basket of four elective courses. Their subject-wise score in those course are given in the dataframe below. Now, we need to make a bar plot, similar to the one we have made previously, showing scores for each student in their selected subjects. As we can see, this dataframe is not tidy — the Students columns has multiple entries for each student. Also, it is not easy to read this table. A better layout for this data would be to have one row per student and have columns corresponding to semester with subjects. In short, a wider format.
Student <- c("Sam","Sam","Sam","Sam",
"Mohan","Mohan","Mohan","Mohan",
"Mike","Mike","Mike","Mike",
"Raj","Raj","Raj","Raj")
Semester <- c("I","I","II","II",
"I","I","II","II",
"I","I","II","II",
"I","I","II","II")
Subject <- c("Maths", "Science","Maths", "Science",
"Maths", "Science","Maths", "Science",
"Maths", "Science","Maths", "Science",
"Maths", "Science","Maths", "Science")
Score <- c(65,75,75,85, 75,85,80,75, 80,90,85,90, 70,80,75,90)
df1 <- data.frame(cbind(Student, Semester, Subject, Score))
df1$Score <- as.integer(df1$Score)
df1 Student Semester Subject Score
1 Sam I Maths 65
2 Sam I Science 75
3 Sam II Maths 75
4 Sam II Science 85
5 Mohan I Maths 75
6 Mohan I Science 85
7 Mohan II Maths 80
8 Mohan II Science 75
9 Mike I Maths 80
10 Mike I Science 90
11 Mike II Maths 85
12 Mike II Science 90
13 Raj I Maths 70
14 Raj I Science 80
15 Raj II Maths 75
16 Raj II Science 90Here, the pivor_wider function creates new columns by combining the values in the Semseter and the Subject columns. Since there are two different values in both these columns so we’ll have four (2x2) new columns. The values for these new columns are taken from the Scores column. We provide custom names to the new columns using the names_glue argument.
df1 <- df1 %>%
pivot_wider(names_from = c(Semester, Subject), values_from = Score, names_glue = "Sem-{Semester}_{Subject}")
df1# A tibble: 4 × 5
Student `Sem-I_Maths` `Sem-I_Science` `Sem-II_Maths` `Sem-II_Science`
<chr> <int> <int> <int> <int>
1 Sam 65 75 75 85
2 Mohan 75 85 80 75
3 Mike 80 90 85 90
4 Raj 70 80 75 90This wider dataframe is not only more readable but also allows to do some quick analysis. For example, the code below calculates the average score per semester for each student.
df2 <- df1 %>%
mutate(SemI_Average = rowMeans(select(df1,contains("Sem-I")))) %>%
mutate(SemII_Average = rowMeans(select(df1,contains("Sem-II")))) %>%
select(Student, contains("Average"), everything())
df2# A tibble: 4 × 7
Student SemI_Average SemII_Average `Sem-I_Maths` `Sem-I_Science`
<chr> <dbl> <dbl> <int> <int>
1 Sam 75 80 65 75
2 Mohan 78.8 77.5 75 85
3 Mike 86.2 87.5 80 90
4 Raj 78.8 82.5 70 80
# ℹ 2 more variables: `Sem-II_Maths` <int>, `Sem-II_Science` <int>Learning how to format the data in the correct format is inarguably in an important skill to have for every data scientist. Once the data is arranged in the required format, we’ll can easily apply different functions to analyze the data and get answers for your questions.